UMD Researchers Discover a Way That Animals Keep Their Cells Identical
Collaboration between biologists and computer scientists yields insights into some cancers and age-related diseases
Cancers arise in skin, muscle, liver or other types of tissue when one cell becomes different from its neighbors. Although biologists have learned a lot about how tissues form during development, very little is known about how two cells of the same tissue stay identical for an animal’s entire lifetime.
A University of Maryland research team is the first to discover that a regulatory protein named ERI-1 helps ensure that all cells in a tissue remain identical to one another. The work involved collaboration between developmental biologists and computer scientists, with the latter contributing their expertise with machine learning analysis. The finding could bring biologists one step closer to understanding some cancers and other age-related diseases.
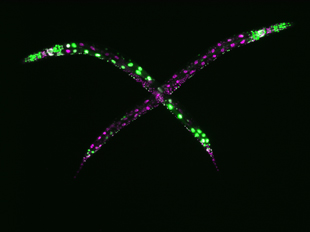 The study, which is the first of its kind conducted in a whole animal (the roundworm Caenorhabditis elegans) instead of cultured cells, appears in the August 1, 2016 issue of the Journal of Cell Biology. The researchers’ approach reveals one important mechanism that animals use to maintain uniform patterns of gene expression. The team’s use of machine learning software proved essential for quickly and clearly identifying complex patterns in the data.
The study, which is the first of its kind conducted in a whole animal (the roundworm Caenorhabditis elegans) instead of cultured cells, appears in the August 1, 2016 issue of the Journal of Cell Biology. The researchers’ approach reveals one important mechanism that animals use to maintain uniform patterns of gene expression. The team’s use of machine learning software proved essential for quickly and clearly identifying complex patterns in the data.
“Cells can look the same and behave the same, but how? The liver is full of liver cells and doesn’t have any heart cells, for example. There’s so much that needs to happen to maintain a tissue,” said Antony Jose, an assistant professor in the UMD Department of Cell Biology and Molecular Genetics and senior author on the study. “It’s a fundamental question that’s been hiding in plain sight. We’ve now proposed an answer that could help advance our understanding of age-related diseases.”
The results suggest that long sections of repetitive DNA can be read differently from cell to cell. The researchers found that, in healthy tissues, ERI-1 normalizes these differences by ensuring that each cell expresses their genes at the same levels. When the researchers turned off the gene that produces ERI-1 in C. elegans, an abnormal patchwork of gene expression appeared in the worms’ intestines.
“To understand these processes, we needed to measure single-cell differences in a whole animal,” Jose said. “We had to know which cell was related to which others and simultaneously measure various properties in all cells within a tissue. Technically, achieving this is very difficult. But you can’t adequately answer these questions outside the context of the whole animal.”
To achieve this complex analysis, Jose and his colleagues formed an unexpected collaboration. Lead author Hai Le (B.S. ’13, biological science), an undergraduate researcher in Jose’s lab who is now a student at the Johns Hopkins University School of Medicine, presented a poster at UMD’s Bioscience Day conference in 2012. Future collaborator Michael Bloodgood, an associate research scientist at UMD’s Center for Advanced Study of Language, stopped by to discuss Le’s work. The two researchers quickly recognized the potential for machine learning to help facilitate Le and Jose’s analysis.
“Linguists use machine learning to compare blocks of text to identify nouns and verbs, analyze sentence structures and determine average word length, for example,” Jose said. “Hai and Michael recognized that we could use the same technique to analyze gene expression in intestinal cells.”
Machine learning software can reveal complex patterns that the human eye cannot see. As the name implies, the software can be taught to look for specific patterns and can also “learn” from experience, becoming more efficient with each subsequent analysis. Using this approach enabled the researchers to quickly make objective comparisons that would have been all but impossible using other methods.
The researchers chose C. elegans because it is a simple organism that can easily be studied at the level of a single cell while it is still alive. Jose notes that their technique is broadly applicable, and could be modified to work with other genes and different tissues as well. If others adopt the team’s whole-animal methodology, Jose believes it could signal a shift in the way cell biologists approach their experimental design.
“The effect of cancer drugs is often examined in cultured cells. Our work suggests that studies on cells outside an animal could miss many things. For example, cultured cells can show differences in gene expression that are eliminated in a whole animal,” Jose said. “I believe our results could lead to some shifts in thinking about how to imitate a whole animal in cell culture conditions.”
###
In addition to Jose, Le and Bloodgood, authors on the study include Monika Looney (B.S. ’16, biological science, psychology) and Benjamin Strauss (B.S. ’12, computer engineering) who were undergraduate researchers when the study was conducted.
The research paper, “Tissue homogeneity requires inhibition of unequal gene silencing during development,” Hai Le, Monika Looney, Benjamin Strauss, Michael Bloodgood and Antony Jose, appears in the August 1, 2016 print edition of the Journal of Cell Biology.
This work was supported by the Howard Hughes Medical Institute and the National Institutes of Health (Award Nos. R00GM085200 and R01GM111457). The content of this article does not necessarily reflect the views of these organizations.