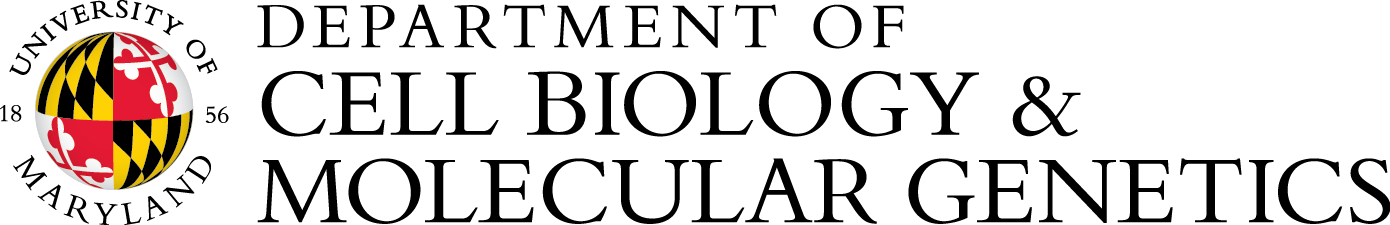Contact Info
Office: 2136 Bioscience Research Bldg
Lab: 2205 Bioscience Research Bldg
Office Phone: 301-405-7028
Lab Phone: 301-405-9513
Antony M. Jose
Professor
Teaching
- Systems View of Cell Biology (BSCI432)
- Quantitative Modeling for Experimental Biologists (CBMG626)
Graduate Program Affiliations
- BISI-Computational Biology, Bioinformatics, & Genomics (CBBG)
- BISI-Molecular & Cellular Biology (MOCB)
Research Interests
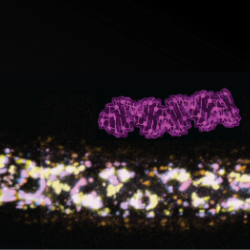
Life relies on regulation from molecular to population scales. We are interested in advancing the understanding of living systems through conceptual and experimental work on regulation. Conceptually, we aim to explore the origins, diversity, and limits of regulatory architectures. Experimentally, we aim to elucidate mechanisms that use RNA to regulate gene expression across space and time using the nematode Caenorhabditis elegans. Please visit our website for more information.
Awards
- 2019 Graduate Faculty Mentor of the Year, UMD
- 2016 National Academy of Sciences Kavli Fellow
Education
- Ph.D., Yale University, 2005
- Postdoc, Harvard University, 2011