Teaching
- Microbial Physiology (BSCI443)
- Gene Expression (CBMG688F)
Graduate Program Affiliations
- BISI-Molecular & Cellular Biology (MOCB)

Research Interests
Protein synthesis is a central process in all cells, and the protein synthesis machinery has been a major target for antibiotics. The rise of multidrug-resistant bacteria and a shortage of new antibiotics demand further studies of the protein synthesis machinery and the mechanisms of antibiotic resistance. Defects during protein synthesis in humans also lead to developmental and neurological diseases, yet the disease-causing mechanisms remain largely unclear. The overall goal of our laboratory is to understand the molecular basis of protein synthesis and its disease connection. We are combining biochemical, single-cell, genetic, genomic, and proteomic approaches to study the following research areas:
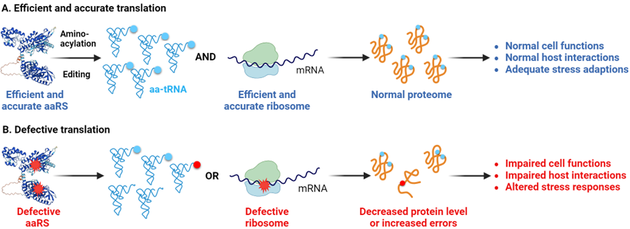
1. The impact of translational defects in bacteria-host Interactions
Protein mistranslation (reduced translational fidelity) has been shown to cause growth defects in bacteria, mitochondrial dysfunction in yeast, and neurodegeneration in mammals. However, the role of translational fidelity during pathogen-host interaction is poorly understood. We have recently shown that optimal translational fidelity is critical for Salmonella to invade host cells. We are currently exploring the mechanisms by which translational fidelity and ribosome assembly affect host interactions, motility, stress responses, and antibiotic tolerance using systems biology and single-cell approaches.
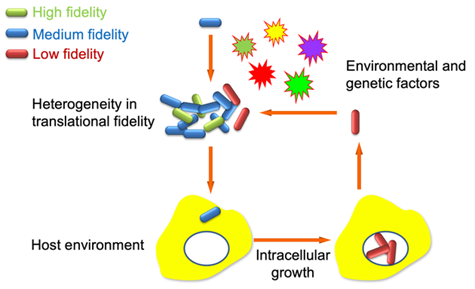
2. Translation and phenotypic heterogeneity among single cells
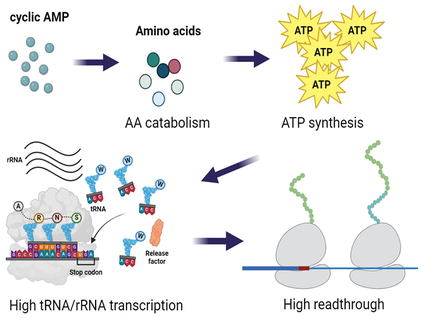
Phenotypic heterogeneity among genetically identical bacterial cells is critical for adaptation to the changing environment and leads to tolerance to antibiotics and stresses. We have shown that translational fidelity and ribosomal RNA expression are heterogeneous and sensitive to the fluctuation of the cellular cyclic AMP level. We are interested in understanding how fluctuations in translation arise and affect phenotypic heterogeneity.
3. Protein Synthesis Defects in Eukaryotes and Human Diseases
Recent advances in genome sequencing and genetics have led to rapid growth of identified mutations in aaRSs and ribosomal proteins that cause human diseases. We are currently using yeast and worm models to understand how translational defects affect stress responses, fitness, and aging.
Education
- Ph.D., The Ohio State University, 2008