New Study Reveals the Final Step in RNA Recycling and Offers Clues for Developing Novel Antibiotics
University of Maryland researchers discovered one enzyme’s vital role in breaking down RNA is more specific than previously thought, revising a vital cellular process misunderstood since the 1960s
New research from the University of Maryland sheds light on the poorly understood process that living cells use to recycle the genetic material they need to survive. The work reveals a previously unknown step in the recycling process that triggered cell death in bacteria when inactivated. Scientists may be able to exploit this trigger and the mechanism behind it to develop new antibiotics. The study was published in journal eLife on June 21, 2019.
"Finding new ways to kill bacteria is becoming more and more difficult as pathogens evolve resistance to our current antibiotics,” said Vincent T. Lee, professor in the UMD Department of Cell Biology and Molecular Genetics and one of the lead authors on the study. “This work opens up opportunities for us to look at a biological process within the cell that we hadn’t known to exist until now.”
RNA Recycling
To understand this new work, it is important to understand what scientists know about RNA and how it is recycled. RNA molecules are long chains of nucleic acids, called nucleotides, which cells use to express the genes in their DNA and generate proteins. Once an RNA molecule has done its job, the cell must break the long chain down into its constituent nucleotides that can be recycled and reused.
The process of breaking down RNA chains, known as RNA degradation, is carried out by a number of enzymes in the cell. These enzymes can be thought of as molecular scissors that come in two varieties: endonucleases, which insert themselves into the middle of long strings of RNA and begin snipping it up into shorter chains; and exonucleases, which grab those shorter chains of RNA from the end and clip off nucleotides one at a time.
Scientists have long known that many different enzymes serve as exonucleases (most bacteria have at least seven). The presumption was that these different enzymes perform overlapping duties, and that the redundancy acts as a sort of fail-safe.
Refining the Final Step
In the new research, Lee and his colleagues showed that there is at least one unique exonuclease whose job is not performed by any other. When a string of RNA has been broken down to just two nucleotides, only one exonuclease can perform the final cut that separates them.
Known as Orn, short for oligoribonuclease, this enzyme is so important that many bacteria—including pathogens such as E. coli, salmonella and Vibrio cholerae—cannot exist without it.
“Scientist have known that Orn was important in the recycling of small chains of RNA, once you got down to seven or 10 nucleotides, since the late 1960s,” Lee said. “But this new work shows that Orn is highly specific to splitting two nucleotides.”
That’s a significant finding, because it changes RNA recycling from a series of activities performed by a variety of generalists to a process that has at least one specialized activity carried out by one specific enzyme. That means there is a vital step that can be targeted by scientists looking for new ways to kill cells.
According to Lee, these findings indicate there is something very significant about chains of just two nucleotides. First, Orn must somehow recognize two nucleotides (known as diribonucleotides) as distinct from RNA chains of three or more nucleotides. Second, the cell must also recognize diribonucleotides, and they must signal something bad, because their persistence somehow causes cells to die.
“This work suggests that the accumulation of diribonucleotides has an effect on cells that we don’t understand,” said Wade Winkler a professor in the UMD Department of Cell Biology and Molecular Genetics and a co-author of the study. “It may be that diribonucleotides serve as a signal that, in some way, induces cell death in these bacteria, which is something that had not been considered before.
In the Lab
To clarify the role of Orn and identify the vital significance of diribonucleotides, the researchers conducted laboratory experiments incubating exonucleases with strands of RNA that varied in length from two to seven nucleotides and observed the reactions over time. Their results showed that when Orn was the only exonuclease present, chains of up to seven nucleotides took significantly longer to degrade than when other exonucleases were present. When other exonucleases were present but Orn was absent, the RNA chains were clipped down to two nucleotides, but never completely reduced to single nucleotides until Orn was added to the medium.
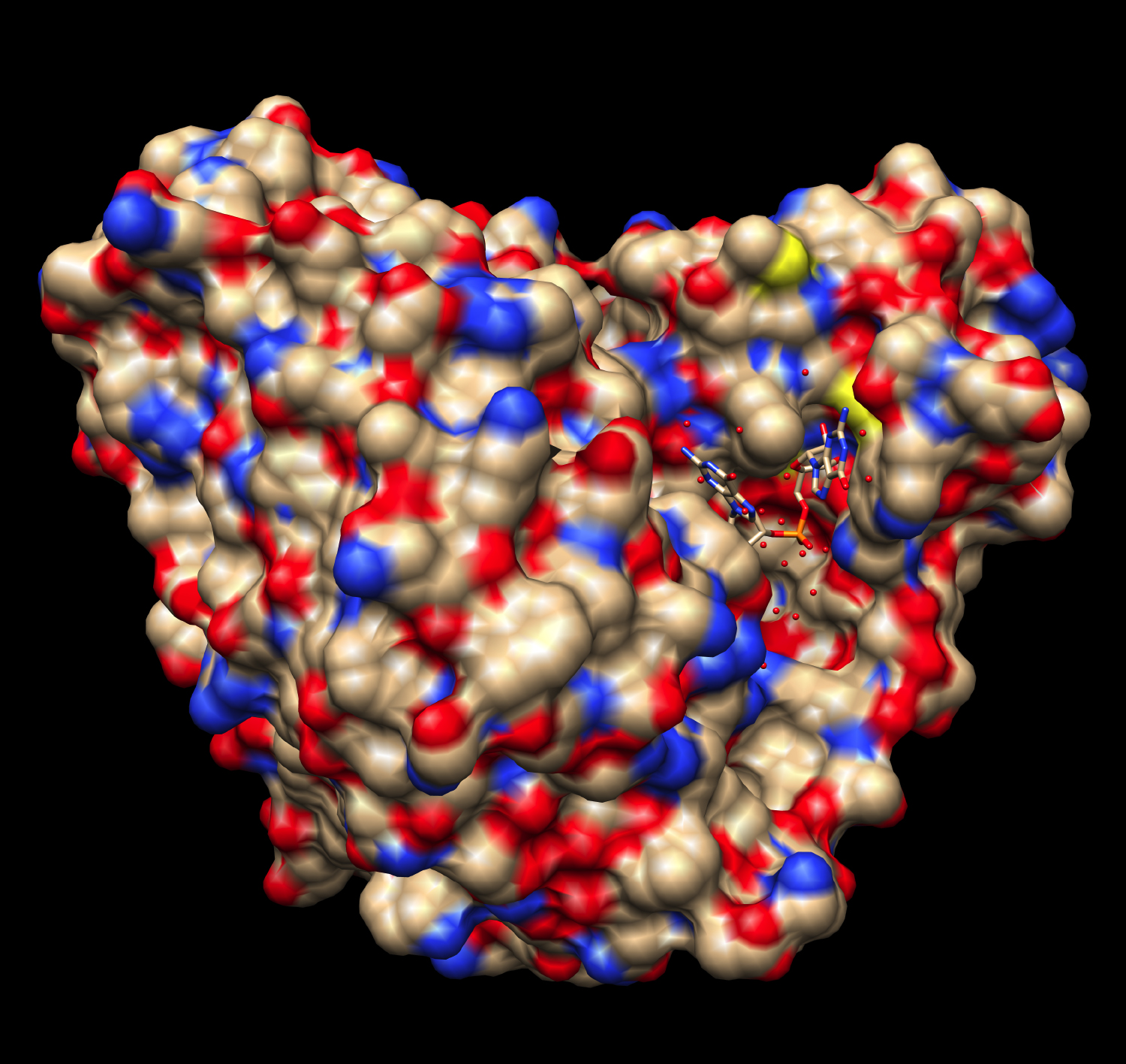
Collaborative work by Lee, Winkler and Holger Sondermann from Cornell University also examined the crystal structures of Orn and found that the active site on the Orn molecule is distinctly suited to a diribonucleotide. The diribonucleotide fits tightly into the folds of the Orn molecule and its precise position allows the bond between nucleotides to be clipped.
A New Role for Orn
Prior to this work, scientists surmised that Orn’s role in cell survival was to ensure maximum recycling of the raw material for making new RNA chains. The assumption was that cells died without Orn because of a bottleneck in recycling that prevented enough raw material from being freed up to make new RNA.
This new work dismisses that assumption. Because Orn selectively goes after diribonucleotides, the majority of short RNA chains get broken down one nucleotide at a time by other exonucleases. If removing Orn caused recycling to bog down at the point of clipping diribonucleotides into single nucleotides, it would take a long time for a cell to run out of nucleotides. But the absence of Orn does not take a long time to kill cells. Rather, most cells simply don’t survive at all when scientists attempt to remove Orn. According to Lee, this indicates that the very presence of diribonucleotides is what kills the cells.
“Both the structural properties of Orn and our biochemical results really suggest that its role as a general exonuclease should be reconsidered,” Lee said. “Nobody knew that Orn cleaving diribonucleotides could be such an important process to investigate. This opens the door on a new direction for pathogen research.”
Going forward, the researchers will explore the reason that diribonucleatides can signal death in bacterial cells with the hope that these studies may lead to the development of new drugs that target those pathways.
###
This work was supported by the National Institutes of Health (Award Nos. R01GM123609, R01AI110740, R01AI142400, T32-GM080201), GM103485); Cystic Fibrosis Foundation (Award No. LEE16G0); and National Science Foundation (Award Nos. MCB1051440 and DMR-1332208). The content of this article does not necessarily reflect the views of these organizations.
The research paper, “A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation,” Soo-Kyoung Kim, Justin D. Lormand, Cordelia A. Weiss, Karin A. Eger, Husan Turdiev, Asan Turdiev, Wade C. Winkler, Holger Sondermann, Vincent T. Lee, was published in the journal eLife on June 21, 2019.
Writer: Kimbra Cutlip
Media Relations Contact: Abby Robinson, 301-405-5845, abbyr@umd.edu
University of Maryland
College of Computer, Mathematical, and Natural Sciences
2300 Symons Hall
College Park, Md. 20742
www.cmns.umd.edu
@UMDscience
About the College of Computer, Mathematical, and Natural Sciences
The College of Computer, Mathematical, and Natural Sciences at the University of Maryland educates more than 9,000 future scientific leaders in its undergraduate and graduate programs each year. The college's 10 departments and more than a dozen interdisciplinary research centers foster scientific discovery with annual sponsored research funding exceeding $175 million.